Running Head: Variable Identification Through Random Forests
Funding Support: Funding for this work was provided by the National Heart, Lung, and Blood Institute NHLBI: R01 HL 114055. Analyses related to the COPD Genetic Epidemiology (COPDGene®) dataset were supported, in part, by NHLBI: R01 HL089856 and R01 HL089897.
Date of acceptance: October 15, 2015
Abbreviations:chronic obstructive pulmonary disease, COPD; Burden of Obstructive Lung Disease, BOLD; forced expiratory volume in 1 second, FEV1; peak expiratory flow, PEF; forced vital capacity, FVC; positive predictive value, PPV; negative predictive value, NPV; COPD Genetic Epidemiology, COPDGene; 6-minute walk test, 6MWT; American Thoracic Society, ATS; St. George’s Respiratory Questionaire, SGRQ; standard error, SE; short of breath, SOB; National Health and Nutrition Examination Survey, NHANES; out-of-bag, OOB; body mass index, BMI
Citation: Leidy NK, Malley KG, Steenrod AW, et al for the High Risk COPD Screening Group. Insight into best variables for COPD case identification: A random forests analysis. Chronic Obstr Pulm Dis. 2016; 3(1): 406-418. doi: http://doi.org/10.15326/jcopdf.3.1.2015.0144
Introduction
A substantial number of individuals with chronic obstructive pulmonary disease (COPD) are undiagnosed.1 Although patients with mild COPD may benefit from treatment, there is little empirical evidence to support this, with the exception of smoking cessation, which should be addressed with all smokers.2 As a result, multiple organizations recommend against screening for asymptomatic COPD.2-5 It is well known, however, that people with moderate to severe airflow obstruction and those at risk for acute exacerbations experience significant health benefits from treatment, including pharmacotherapy and rehabilitation.6 Identifying and treating these individuals should lead to better outcomes at the patient, practice, and population levels.7
Spirometry is the gold standard for confirmation of a COPD diagnosis3 and has been used to screen high-risk patients in pulmonary clinics.8 Rigorous administration of this test by trained personnel to all patients in primary care settings can be difficult and expensive, with cost-effectiveness a concern when the yield may be 10% to 50%, depending on the setting, half of whom likely have mild disease.2,9-14 Questionnaire-based screening offers a practical method for identifying people who may have clinically significant COPD. Including peak expiratory flow (PEF) in the screening process could enhance efficiency by reducing the number of false positives.
To date, questionnaires have been designed to identify people with COPD (forced expiratory volume in 1 second [FEV1]/forced vital capacity [FVC] ratio <0.70) without reference to disease severity or exacerbation risk.15-22 The ability of these tools to detect cases have been modest,2 with sensitivity/specificity ranging 66% / 54% for an 8-item diagnostic questionnaire tested in the general population23 to 87% / 71% for a 6-item questionnaire in primary care,15 the latter associated with a positive predictive value (PPV) of 38% and a negative predictive value (NPV) of 97%. Nelson et al24 tested a three-staged approach (questionnaire, PEF, and spirometry) for identifying moderate to severe COPD (FEV1<60% predicted) in the general population. Six percent of 3791 participants (n=227) with 2 or more risk factors had abnormal PEF values, suggesting a more sensitive screening questionnaire is needed to find the more severe cases.
The current study was part of a larger multi-method project to develop a practical and effective primary care strategy for identifying undiagnosed patients with clinically significant COPD, defined as an FEV1 % predicted < 60%, or at risk of developing acute exacerbations. The project began with a comprehensive literature review of screening questionnaires and epidemiological studies of risk factors for acute exacerbations of COPD to identify candidate constructs for the new case-finding tool.25 Qualitative focus groups were conducted to understand how patients describe risk factors and manifestations of COPD, in order to further inform questionnaire content.26
The purpose of this component of the larger project was to examine 3 existing databases for additional empirically-based insight into attributes that characterize COPD and the categories and types of variables that may be useful in case identification. Results were used in conjunction with the literature review25 and qualitative research26 to develop a pool of candidate items for empirical testing.
Methods
This study was a retrospective analysis of 3 existing and available databases: COPD Foundation PEF study24 (N=5761), the Burden of Obstructive Lung Disease (BOLD) study, Kentucky site27 (N=508) and COPD Genetic Epidemiology (COPDGene®) (Supported by NHLBI R01 HL089856 and R01 HL089897)28 (N=10,214). Each was a prospective study conducted in the United States, enrolling a convenience sample of COPD and non-COPD participants. Characteristics of each dataset are shown in Table 1. No study was specifically designed to identify cases of COPD, but each included samples and variables suitable for case or control assignment and comparisons.
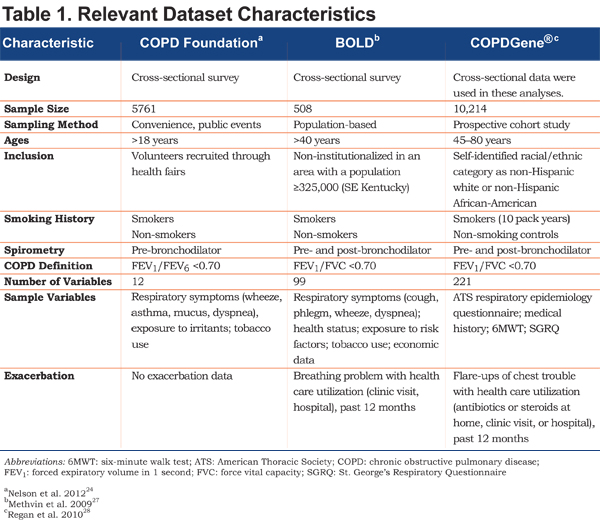
Random forests was used to analyze the data. Briefly, random forests is a machine learning statistical method that uses decision trees to identify and validate variables most important in prediction29; in this case, classifying or predicting group membership in each of 4 case-control scenarios. Decision trees for group membership are constructed with randomly selected subsets of participants and variables. Forests of these decision trees are built, which together make a prediction for each participant. These results are used to identify and validate variables most important to the prediction. Estimates of sensitivity, specificity, and overall error rate are computed to indicate how well the variable sets predict cases and controls. Random forests and other machines are not based on models, and therefore avoid problems of model misspecification or invalid assumptions. Additional information on random forests is provided in the online supplement.
Case-control Definitions and Variable Selection
Four case-control scenarios were tested, as permitted by the available data (see Table 2). For each scenario, random forests were used to identify the best set of variables that could differentiate cases and controls. Scenario 1 was designed to identify variables that best differentiate COPD patients with moderate to severe airflow limitation (FEV1 less than 50% predicted3) (cases) from those without COPD (controls). Because these 2 groups represented extremes from an airflow obstruction perspective, they should, theoretically, be relatively easy to differentiate, with the smallest number of variables and the lowest error rates. This provided context for interpreting the remaining scenarios and demonstrated the presence of a detectable signal in the 3 datasets. The purpose of Scenario 2 was to identify variables that distinguish undiagnosed and diagnosed COPD, providing insight into patient attributes associated with a missed diagnosis. Scenario 3 differentiated COPD patients with an exacerbation history (cases) and those without exacerbation history (controls), to determine what attributes may be unique to this specific high-risk group. Finally, Scenario 4 identified attributes differentiating patients with an FEV1 <60% or an exacerbation history from all others, including COPD with higher FEV1 % predicted and no exacerbation and non-COPD patients, replicating the purpose of the new screening tool. Based on data availability for group classification, Scenarios 2 and 3 were tested with data from the BOLD and COPDGene® datasets and Scenario 4 was tested only in the COPDGene® dataset.
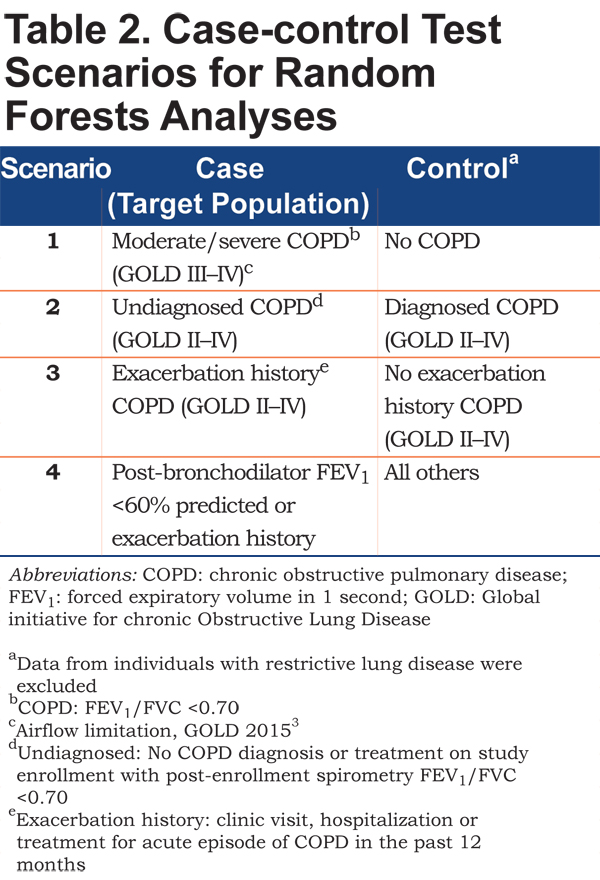
All clinical, demographic, and patient-reported variables comprising the dataset were used in the analyses, with 2 exceptions. First, to avoid circular reasoning, variables synonymous with case definitions of COPD (e.g., spirometry or record of maintenance therapy) or COPD exacerbation (treatment with antibiotics, steroids, or COPD hospitalization) were excluded. Second, to facilitate interpretation and instrument development, questionnaire subscale or total scores were excluded (individual questionnaire items were included). The number of candidate variables in each dataset is shown in Table 3.
Analyses
Statistical
The goal was to derive the smallest set of predictor variables that could differentiate cases and controls with a degree of accuracy (error rate) comparable to larger sets of variables. For each scenario, the first random forests analysis was performed with all variables in the dataset, with the exceptions outlined above. The variable importance measure was used to remove the least important variables and new random forests analyses were performed, keeping the final error rate in the same range as the original all-variable error rate. Variable importance is the mean decrease in prediction accuracy when the variable’s values are randomly permuted, standardized to a 0–100 range with higher values indicating greater relative importance. This rating is a function of all other variables in the model; if one or more variables are removed, the importance rating changes.
The number of predictor variables was not reduced if the reduction caused more than a 2%–3% increase in the out-of-bag (OOB) error rate for the analysis. The OOB error rate is the misclassification rate resulting from each tree being tested on data not used to build the tree (the OOB sample), averaged over all trees in the forest and then over all forests in the analysis. With the best sets identified, sensitivity and specificity of each set were computed, where sensitivity is 1 – (error rate for cases) and specificity is 1 – (error rate for controls).
The R package randomForest was used to perform the analyses.30 Additional information on the use of random forests in this study is provided in the online supplement.
Thematic
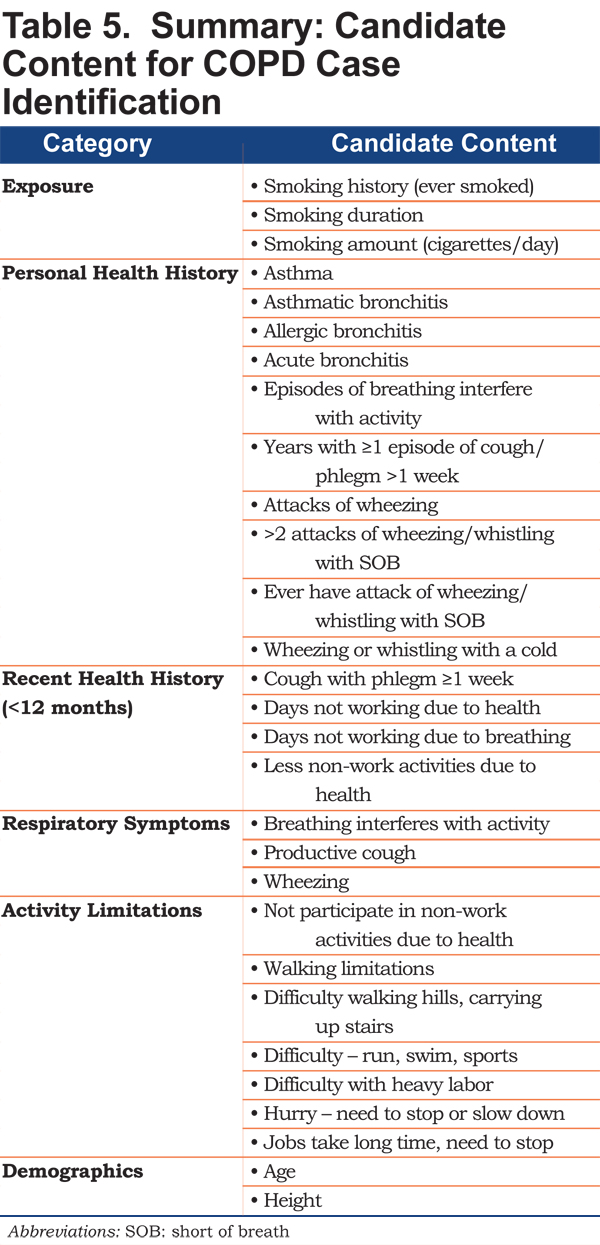
Variables that emerged as important within and across the 4 case-control scenarios were organized by theme using the classification system derived through the literature review25 and qualitative research.26 Specifically, each variable was assigned to 1 of 6 categories of variables that could be useful for identifying undiagnosed cases of COPD: exposure, personal health history, recent health history, respiratory symptoms, activity limitations, and demographics. These variables would be examined together with information from the literature and qualitative research to develop candidate items for the new questionnaire.
Results
Number of Variables and Out-of-Bag Error Rates
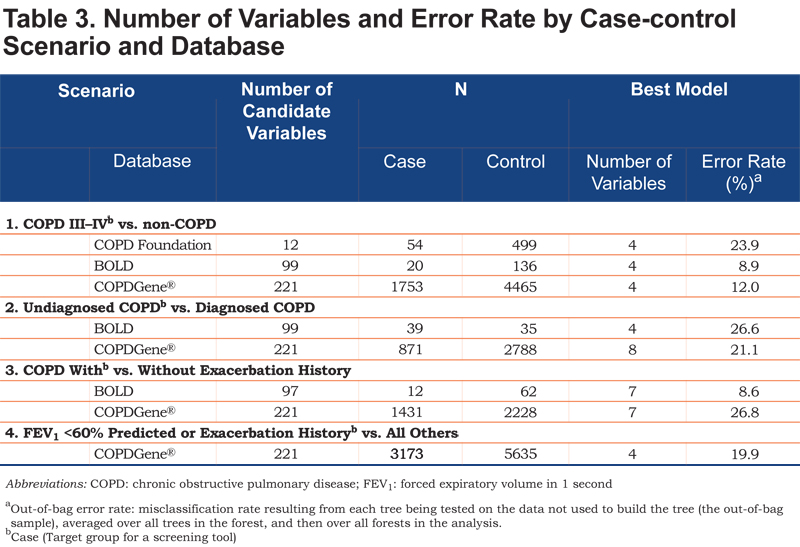
Table 3 summarizes the number of candidate variables, sample sizes, and the number of variables and error rates associated with the best models, stratified by scenario and database. Each scenario identified small sets of variables with the best predictive ability from the full list of candidate variables; as few as 4 variables were able to differentiate cases and controls.
In the first case-control scenario, each model included 4 variables that best differentiated cases and controls, with OOB (misclassification) error rates of 9% (BOLD), 12% (COPDGene®), and 24% (COPD Foundation). Overall, the error rates for the second case-finding scenario were higher, indicating it was more difficult to differentiate undiagnosed and diagnosed cases of COPD. The third case-finding scenario produced 7-variable models, with greater predictive accuracy (lower error rate) in the BOLD dataset (9%) relative to the COPDGene® (27%). In the final scenario, 4 variables emerged from the 221 candidate variables, with an error rate of 20%.
Variables, Importance Indicators, Sensitivity and Specificity
For each case-control scenario and dataset, the most important variables, their importance ratings, and the sensitivity and specificity of the variable set are shown in Table 4.
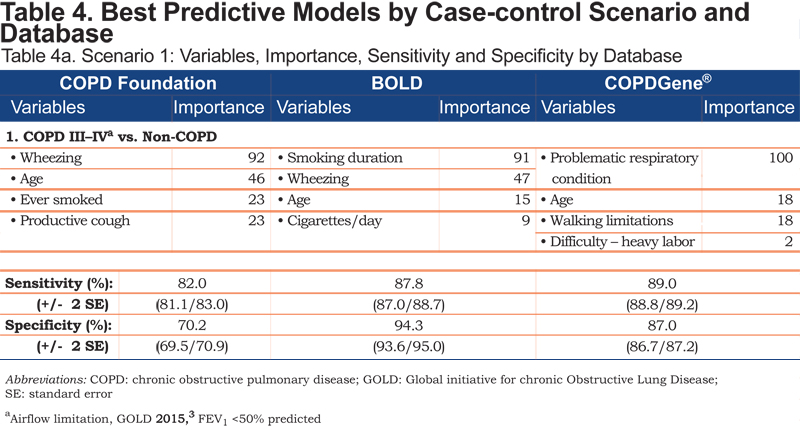
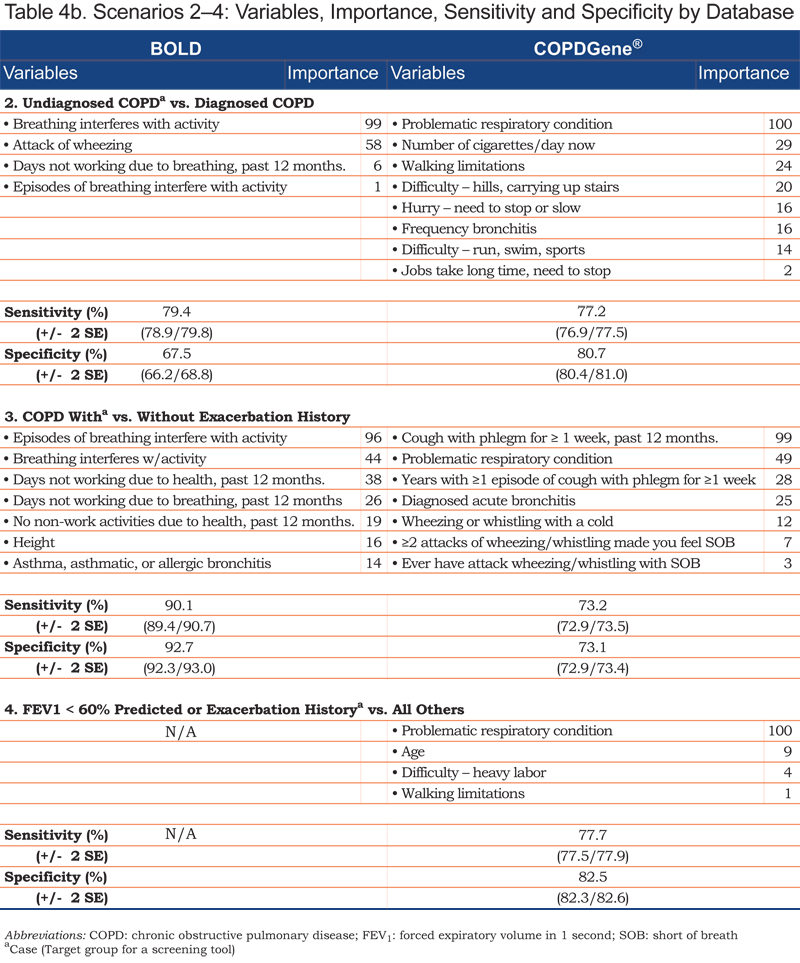
Age emerged in each of the Scenario 1 models; smoking history and wheezing were contributing variables in the COPD Foundation and BOLD datasets. Individual variables differentiating undiagnosed from diagnosed COPD included breathing-related or general activity limitations. COPD patients with and without exacerbation history (Scenario 3) were distinguished by reports of episodic breathing-related issues, including recent (past 12 months) history of cough with phlegm for more than a week and/or missed work days and non-work activities. General history of episodes of breathlessness interfering with activity; acute wheezing with shortness of breath; acute bronchitis; wheezing with a cold; or a history of asthma, asthmatic or allergic bronchitis were also differentiating variables for this scenario.
Each set of variables for any given scenario included 1 or 2 predictors that played the greatest role in determining the outcome or correct classification of a participant within the model. Smoking duration and wheezing were key variables for differentiating moderate to severe COPD from non-COPD patients in the BOLD and COPD Foundation datasets. In the COPDGene® dataset, patient self-rating of their respiratory condition was important in all 4 case-finding scenarios. Variables that emerged across multiple case-finding scenarios included patient report of walking limitation due to shortness of breath (COPDGene®), breathing problems interfering with activity (BOLD), and missed work and non-work activity in the prior 12 months (BOLD).
Thematic Summary
The variables identified in the 4 case-control scenarios organized into 6 categories are shown in Table 5. Personal health history variables differentiating cases and controls included asthmatic, allergic, or acute bronchitis and episodes of wheezing; current respiratory symptoms included reference to breathing interfering with activity, productive cough, and wheezing. Variables related to activity limitations included difficulty with moderate to strenuous activity.
Discussion
The purpose of this study was to explore 3 existing databases to uncover variables that may be useful in the identification of patients with undiagnosed clinically significant COPD. The intent was to synthesize this information with insight from the literature25 and qualitative research26 to develop a pool of candidate items for a new screening questionnaire, ready for quantitative testing.
Screening questionnaires should be short, easy to administer, and simple to score, with a balance of sensitivity and specificity that engenders clinical interest and confidence.31,32 Higher levels of sensitivity will permit fewer missed patients, with the added costs of spirometric testing in people without clinically significant COPD; greater specificity will result in more missed cases, but fewer false positives and lower overall screening costs.33 These analyses attempted to identify the best and smallest set of predictors capable of differentiating cases and controls under 4 scenarios, optimizing the balance between number of variables and precision.
Across scenarios and datasets, as few as 4 to 8 variables, from a starting set of 12 to 221 candidate variables, were able to differentiate cases and controls, with error rates of 9% to 27%. Sensitivities/specificities ranged from 79% / 68% for under diagnosis to 90% / 93% for differentiating COPD patients with and without exacerbation history, both in the BOLD dataset. This suggests a short screening questionnaire of 4 to 8 carefully selected items is a feasible and reasonable objective for a new, targeted screener. Existing screeners for uncovering new COPD cases, without reference to severity or exacerbation risk, range from 3 items, with a sensitivity of 78% and specificity of 65% in the general population21 to 10 items, with a sensitivity of 71% and specificity of 62% in primary care.16
As expected, differentiating moderate to severe COPD cases from non-COPD controls (Scenario 1) was “easiest,” with random forests uncovering 4 variables capable of distinguishing these groups with relatively little error. Age was a consistent variable across the datasets; smoking history and wheeze appeared in 2 of the models. Analyses of the COPDGene® dataset, the largest with respect to the number of variables and sample size, showed that walking limitations and difficulty with heavy labor together with age and respondent perception of a problematic respiratory condition formed the best variable set (12% error rate; sensitivity/specificity = 89% / 87%). This same set distinguished the clinically significant COPD cases from all others (Scenario 4), although the higher error rate (20%;sensitivity/specificity=78% / 82%) suggests this specific target group may be more challenging to identify, particularly when participants with mild COPD are considered controls.
Variable sets capable of identifying cases with an exacerbation history (Scenario 3) included individual variables capturing episodes or “attacks” of shortness of breath; cough with phlegm, or wheezing/whistling; or a diagnosis of acute bronchitis. These results provide insight into the types of questions that could be asked of people without a diagnosis of COPD to uncover new cases at risk of future exacerbations. This assumes evidence suggesting exacerbation history is an important predictor of future exacerbations34 holds true for these individuals as well. It is noteworthy that age, smoking, and walking limitations did not appear in these variable sets.
The purpose of Scenario 2 analyses (undiagnosed versus diagnosed COPD) was to see if there were any defining features that differentiate patients with undiagnosed versus diagnosed COPD. Undiagnosed individuals were those with spirometry indicating the presence of COPD, but no reported diagnosis or treatment. Current smoking appeared in the COPDGene® set, but not in the BOLD. Age, cough, and phlegm were noticeably absent, indicating these are not differentiating features of diagnostic status. On the other hand, dyspnea surfaced in both datasets in the form of activity limitation, which suggests that dyspnea-related questions framed in terms of impact (i.e., hurrying, climbing hills and stairs, or engaging in activity or sports) may be useful for identifying patients with respiratory-related impairment. This is particularly important, given current recommendations that patients who do not recognize or report respiratory symptoms are not targets for screening.2 Helping patients recognize breathing-related impairment may be key to finding individuals most likely to benefit from treatment.
It is important to note that the results are limited by the available data, including samples, settings, and variables. The fact that smoking was the only variable that emerged in the exposure category, for example, is a function of dataset characteristics, e.g., few exposure questions asked and United States study populations with presumably low incidence of biomass fuel exposure. Further, the research settings were varied and not specifically primary care. Additional limitations of this work were the use of self-reported diagnoses for identifying COPD cases and the designation of undiagnosed COPD (Scenario 2) based on spirometry/airflow limitation. Datasets with clinician-confirmed cases of COPD may have yielded more precise predictive models and/or different variable sets. Finally, these analyses were designed to uncover variables for identifying patients with clinically significant COPD and did not test for variables that may uncover mild cases.
Results offer insight into the types of variables that should be considered in developing a new instrument for identifying undiagnosed cases of clinically significant COPD, complementing and extending results of the literature review25 and qualitative research.26 Existing screening measures cover some of the content identified here, to varying degrees. Several symptom-based screening questionnaires, for example, include cough, phlegm, dyspnea, and wheeze,15,19,35,36 while others include personal history of chest infections and breathing-related disability or hospitalizations.20,35 Few questionnaires ask both symptom and exacerbation-related questions.35 The literature review25 supported the exposure/smoking history, personal health history, respiratory symptoms, and impact categories, and identified allergies and body mass index (BMI) as candidate items. The epidemiologic literature review also identified family history, childhood illness, frequency of primary care visits, and fatigue/tiredness as potentially useful variables.25 The qualitative data uncovered additional content, including exposure to second hand smoke and “dirty air,” and non-respiratory symptoms such as lack of energy, sleep difficulties, or slowing down. 26 This information was synthesized to develop a pool of candidate items covering 6 categories of information (exposure, family and personal history, recent respiratory history, respiratory symptoms, non-respiratory symptoms, and impact) ready for quantitative testing in a separate, prospective, case-control study.
Conclusion
Although several screening tools are available to identify patients with undiagnosed COPD, there are no instruments for identifying those most likely to benefit from treatment, i.e., people with moderate to severe disease or at risk of exacerbation. This study was part of a larger project to develop an efficient screening strategy for identifying these patients in primary care. Data from 3 existing COPD databases were analyzed to gain insight into the number and types of demographic and clinical variables that should be considered during questionnaire development. Results were examined with information from the literature and qualitative research to develop a pool of candidate questions ready for empirical testing.
Acknowledgements
The authors thank Kathryn Miller of Evidera for text editing and formatting.
Declaration of Interest
Dr. Leidy, Ms. Steenrod, and Dr. Bacci are employees of Evidera, a health care research firm that provides consulting and other research services to pharmaceutical, device, government, and non-government organizations. In this salaried position, they work with a variety of companies and organizations and receive no payment or honoraria directly from these organizations for services rendered. Ms. Malley is a non-salaried employee of Evidera, and a salaried employee of Malley Research Programming, Inc. In the latter capacity, she provides custom computer programming services to contract research organizations.Dr. Mannino has received honoraria/consulting fees and served on speaker bureaus for GlaxoSmithKline PLC, Novartis Pharmaceuticals, Pfizer Inc., Boehringer-Ingelheim, AstraZeneca PLC, Forest Laboratories Inc., Merck, Amgen, and Creative Educational Concepts. Furthermore, he has received royalties from UptoDate and is on the Board of Directors of the COPD Foundation. Dr. Make has participated in research studies and/or served on medical advisory boards for AstraZeneca, Boehringer-Ingelheim, CSL Bering, GlaxoSmithKline, Forest, Novartis, Spiration, and Sunovion. Dr. Bowler's work has been funded by the National Institutes of Health, Flight Attendant Medical Research Institute, Butcher Foundation, and John W. Carson Foundation. He participates in AstraZeneca and GlaxoSmithKline sponsored clinical trials. He has received compensation as a member of scientific advisory boards of Boehringer Ingelheim Pharmaceutical. Dr. Thomashow has consulted for Boehringer-Ingelheim and has been on advisory boards for GlaxoSmithKline PLC, Novartis, AstraZeneca PLC, and Forest. Dr. Barr has received grant funding for this work from the National Heart Lung and Blood Institute under R01HL114055. Dr. Barr has received grant support from the National Institutes of Health, the United States Environmental Protection Agency, and the Alpha-1 Foundation; he has received royalties from UpToDate. Dr. Rennard was employed by the University of Nebraska Medical Center during the conduct of this study and remains the Richard and Margaret Larson Professor of Pulmonary Research at UNMC and had a number of relationships with companies who provide products and/or services relevant to outpatient management of chronic obstructive pulmonary disease, including A2B Bio, Almirall, APT, AstraZeneca, Boehringer Ingelheim, Chiesi, CME Incite, CSL Behring, Dailchi Sankyo, Decision Resources, Dunn Group, Easton Associates, Forest, Gerson, GlaxoSmithKline, Johnson and Johnson, Medimmune, Novartis, Novis, Nycomed, Otsuka, Pearl, Pfizer, PriMed, Pulmatrix, Roche, Takeda, Theravance; these relationships include serving as a consultant, advising regarding clinical trials, speaking at continuing medical education programs and performing funded research both at basic and clinical levels. Dr. Rennard is currently employed by AstraZeneca in which he owns shares. He does not own any stock in any other pharmaceutical companies. Dr. Houfek declares no conflict of interest. Dr. Yawn has received research funding from the National Institutes of Health, Agency for Healthcare Research and Quality, the Centers for Disease Control and Prevention, and from Boehringer Ingelheim for research on COPD. Dr. Yawn has received compensation from Merck and Forrest for COPD advisory boards, and Grifols for an advisory board on alpha-1 antitrypsin deficiency states. Dr. Han has consulted for GlaxoSmithKline, Boehringer-Ingelheim, and Regeneron. She has served on speaker bureaus for GlaxoSmithKline, Novartis, Boehringer-Ingelheim, Forest, and Grifols. Dr. Meldrum declares no conflict of interest. John W. Walsh declares no conflict of interest. Dr. Martinez has participated in steering committees in COPD or idiopathic pulmonary fibrosis sponsored by Bayer, Centocor, Forest, Gilead, Janssens, GlaxoSmithKline, Nycomed/Takeda and Promedior. He has participated in advisory boards for COPD or idiopathic pulmonary fibrosis for Actelion, Amgen, Astra Zeneca, Boehringer Ingelheim, Carden Jennings, CSA Medixcal, Ikaria, Forest, Genentech, GSK, Janssens, Merck, Pearl, Nycomed/Takeda, Pfizer, Roche, Sudler & Hennessey, Veracyte, and Vertex. He has prepared or presented continuing medical presentations in COPD or idiopathic pulmonary fibrosis for the American College of Chest Physicians, the American Thoracic Society, CME Incite, Center for Health Care Education, Inova Health Systems, MedScape, Miller Medical, National Association for Continuing Education, Paradigm, Peer Voice, Projects in Knowledge, Spectrum Health System, St. John’s Hospital, St. Mary’s Hospital, University of Illinois-Chicago, University of Texas Southwestern, University of Virginia, UpToDate, and Wayne State University. Dr. Martinez has participated in data safety monitoring committees sponsored by GlaxoSmithKline and Stromedix. He has aided with Food and Drug Administration presentations sponsored by Boehringer Ingelheim, GlaxoSmithKline, and Ikaria. He has spoken on COPD for Bayer, Forest, GlaxoSmithKline, and Nycomed/Takeda. He has participated in advisory teleconferences sponsored by the American Institute for Research, Axon, Grey Healthcare, Johnson & Johnson, and Merion. He has received book royalties from Informa.