Running Head: COPD9USA CME Summary: Human Lung Microbiome
Abbreviations: chronic obstructive pulmonary disease, COPD; bronchoalveolar lavage, BAL; Global initiative for chronic Obstructive Lung Disease, GOLD; human immunodeficiency virus, HIV; pneumocystis carinii pneumonia, PCP; cystic fibrosis, CF
Citation: Gates K, Martinez F. The human microbiome in the lung. Chronic Obstr Pulm Dis. 2016; 3(1): 466-472. doi: http://doi.org/10.15326/jcopdf.3.1.2015.0174
Introduction
The lower respiratory tract has historically been considered a sterile environment in healthy individuals.1 Recently, however, there have been several studies suggesting that the lower respiratory tracts of healthy persons are not sterile and have many different bacteria that comprise the lung microbiome.2,3 The lung microbiome is determined by 3 factors: microbial immigration, regional growth conditions, and microbial elimination.2 The balance of these factors is proposed to be one of the key determinants of the healthy lung, as well as having a role in the development and/or progression of chronic lung diseases, including chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis. Given increasing recognition of the existence of the healthy human microbiome,it has become very important to understand the lung microbiome and its role in lung homeostasis and the development of lung disease.4 The “Human Microbiome in the Lung” session during the COPD9USA conference explored the role of the lung microbiome, including bacteria, viruses, and fungi, in COPD.
The Way Forward
In efforts to explore and understand the lung microbiome, the use of newer technologies for culture-independent analysis of bacteria, including high-throughput sequencing of bacterial DNA, has become increasingly utilized. The 16S rRNA gene is a small and highly conserved locus of the bacterial genome which is easily sequenced and can be used to isolate thousands of genome sequences present in a single respiratory sample.2 Using bioinformatics software and taxonomic databases, a population census can be determined for individual specimens allowing the evaluation of individual specimens including determination of total bacterial abundance (the number of times a taxonomic group is counted in a specimen), relative abundance (identification of a predominant taxonomic group in a specimen), and community features (the diversity of taxonomic groups in a single specimen).
16S rRNA sequencing is being used to explore many questions regarding the human lung microbiome, including evaluating the validity of bronchoscopy as a technique to evaluate the bacterial communities of the lower respiratory tract. Bronchoscopy entails insertion of a bronchoscope through the upper airway, including the nose or mouth, to gain access to the lower respiratory tract for sampling. There is a theoretical risk of contamination of the lower respiratory tract with microbes from the upper tract during this procedure. There have been several studies evaluating this risk including Bassis et al.5 In this study, samples from 12 healthy volunteers were obtained from varying locations throughout the respiratory tract including the mouth, lung, gastric aspirate, and nasal cavity. 16S rRNA was detected in all samples, including bronchoalveolar lavage (BAL) fluid, supporting the hypothesis that the lung is not a sterile environment (Figure 1). Interestingly, the 16S rRNA copies were 100 to 1000-fold lower in the BAL samples compared to those of the oral and gastric samples. Further analysis of these samples showed that bacterial composition of the BAL fluid significantly differed from the oral, gastric, and nasal microbiome, supporting the presence of a unique lung microbiome in the lower respiratory tract of healthy individuals. This analysis, along with others, supports the concept that the lower respiratory tract bacterial community closely resembles the bacterial communities of the stomach and the mouth, while differing significantly, suggesting that the oral microbiome is the primary source of bacteria in the lung microbiome.
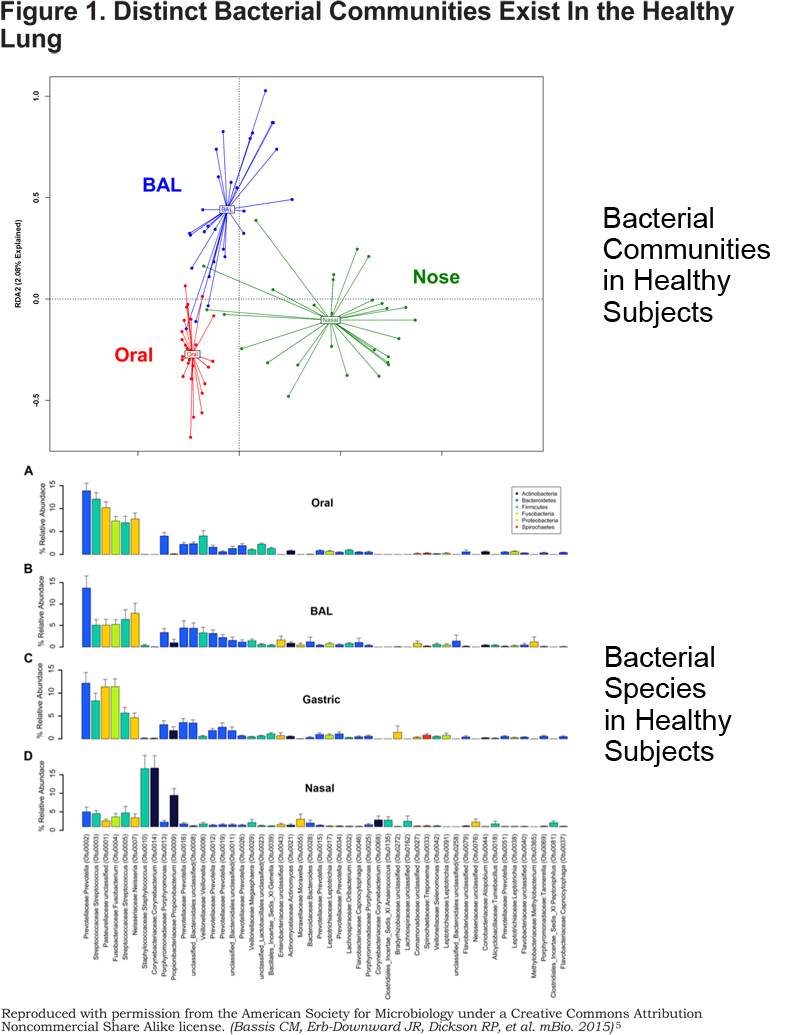
Pulmonary anatomy has also been considered a contributing factor to bacterial communities in the lung microbiome. However, Dickson et al reported there is less spatial variation in the healthy lung microbiota within an individual person, but showed that it does vary between individuals.6 This study also supported the observation that bronchoscopy for evaluation of lavage fluid is an adequate sampling of the lower respiratory tract, with minimal evidence of contamination of the lower respiratory tract as a result of bronchoscopy.
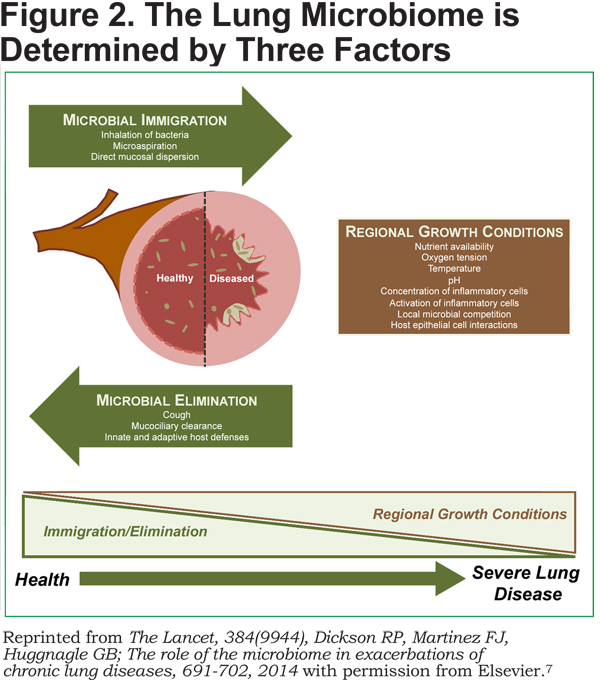
Based on these bacterial community studies, it has been hypothesized that with the development of severe lung disease, there are alterations in regional growth conditions that support changes in the healthy lung microbiota that may lead to the development of chronic lung diseases (Figure 2).7
COPD: Is it Just Bacteria?
COPD is a heterogeneous disease that is characterized by chronic lung inflammation and incomplete reversible airflow limitation. Many COPD patients have frequent exacerbations, often thought to be triggered by an infectious source. Bacteria have historically been associated with COPD exacerbations and progression of disease. Approximately 50% of COPD exacerbations have been associated with bacterial infections including Streptococcus pneumonia, Haemophilus influenza, and Moraxella catarrhalis.8 Whether these organisms represent new infections or simply reflect the lung microbiome in this patient population is an area of continued research. It is thought that the host immune response to bacteria of the lung microbiome may contribute to the pathogenesis of COPD via chronic inflammation.9 However, there are many other microbes including viruses (virome) and fungi (mycobiome) that may contribute to the lung microbiota and the pathogenesis of COPD through initiation of chronic inflammation that may lead to lung injury directly or through changes in the bacterial community of these patients.
Molyneaux et al performed a study to evaluate the effect of pulmonary viral infections on the bacterial airway microbiome in patients with COPD.10 In this study, patients with mild COPD (Global initiative for chronic Obstructive Lung Disease [GOLD] Stage II disease) were compared to healthy controls. The study participants were inoculated with a low dose of rhinovirus and induced sputa were collected prior to rhinovirus infection and at several time points after infection. Gene sequencing of the 16S rRNA gene in sputum samples was performed. Prior to infection there were no significant differences in bacterial burden between COPD patients and healthy controls. However, there was a 6-fold increase in the bacterial load of COPD patients after rhinovirus infection with minimal changes observed in control samples. Additionally, there was a positive correlation between sputum inflammatory markers and bacterial load in the COPD group. In addition to the changes in the number of bacteria, there were also changes observed in the bacterial phyla of COPD patients, such as an increase in the abundance of Proteobacteria, primarily H. flu. The results of this study suggest that viral pulmonary infections may lead to changes in the lung microbiome and may be a cause of secondary bacterial infections in COPD patients.
Fungi are ubiquitous in the environment and are often overlooked due to the difficulty of culturing these organisms with standard techniques. With increasing immunosuppression in COPD patients and increased use of antibiotics, fungal overgrowth is a concern in these patients. Similar to bacteria and viruses, fungi can promote local and systemic inflammation that may contribute to the pathogenesis of COPD. Human immunodeficiency virus (HIV) is an independent risk factor for the development of COPD, though the pathogenesis in this patient population remains unclear. The mycobiome as a potential causative factor in the development of COPD in this patient population has been studied. Cui et al evaluated the mycobiome in BAL fluid obtained from patients with HIV compared to HIV-negative controls.11 HIV-positive patients were noted to have different fungal communities compared to the HIV-negative patients. Differences in fungal communities were also observed in HIV-positive patients with COPD versus similar patients with normal lung function. The predominant difference in the fungal community in this study was due to Pneumocystis jirovecii, an opportunistic infection known to cause pneumonia in HIV positive patients, commonly known as pneumocystis carinii pneumonia (PCP). In patients with HIV, Pneumocystis colonization is associated with an increase in airway obstruction independent of smoking history and is associated with an increase in MMP-12, a protease thought to be important in the pathogenesis of COPD.12 Pneumocystis jirovecii has also been associated with COPD in HIV-negative populations.13 Morris et al showed an association between COPD severity and Pneumocystis colonization, with increased colonization observed in patients with GOLD Stage IV disease. Additionally, animal studies have shown the development of airflow obstruction and emphysema seemingly as a result of Pneumocystis colonization.14,15 Based on these studies, the virome and mycobiome should be further evaluated to better understand their impact on the development and/or disease progression of COPD.
What We Have Learned From Other Disease States and How It Applies to COPD
The lung microbiome has been shown to be important in several lung diseases such as cystic fibrosis (CF). Gaining greater understanding of the role of the lung microbiome in COPD pathogenesis may be better understood by applying information obtained from studies of lung microbiome in CF. Similar to COPD, high throughput techniques have been used to evaluate the complex microbiota of CF revealing similar complexity in the lung microbiome of patients with COPD.16,17 Like CF bacterial communities, COPD bacterial communities demonstrate significant heterogeneity in individual lungs of patients with severe COPD (Figure 3).18
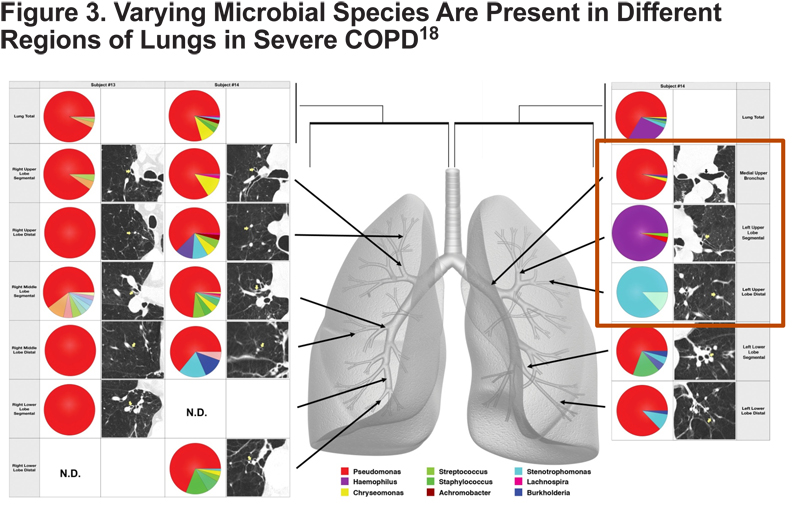
Currently, the role of the microbiome in the pathogenesis of COPD or its role in disease progression is unclear. In efforts to evaluate this further, 16S rRNA sequencing was used to evaluate the bacterial community of patients with severe, GOLD Stage IV COPD.19 There was an association between lung tissue microbiome and changes in both structural and inflammatory markers in the lung tissue. Through many studies the presence of a unique and heterogeneous lung microbiome has been well established, however, understanding the factors that influence the composition of the microbiome and how these alterations may contribute to lung disease requires further exploration in COPD patients. Huang et al evaluated the induced sputum of COPD patients pre- and post-exacerbations showing a change in the microbial composition of the microbiome during exacerbation and with treatment, which included antibiotics and/or steroids.
There is mounting evidence supporting the presence of stable bacterial communities comprising the lung microbiome and the potential role of these organisms in the pathogenesis of COPD. Alterations in these bacterial communities likely play a key role in COPD exacerbations and progression of lung disease with decline in lung function.
Conclusion
The human microbiome, with the emphasis on the upper airway, skin, gastrointestinal tract, and urogenital tract as sources of bacteria contributing to the human microbiome, has been well established.4 There is a growing body of literature suggesting the presence of a lung microbiome as well; representing microbial communities that are unique to the lung and may play a role in the development and/or progression of lung disease. COPD is a chronic lung disease associated with chronic inflammation, airway limitation, and frequent infectious exacerbations that contribute to progression of the disease. Understanding the role of the lung microbiome in COPD and how it may influence pathogenesis and disease progression using new technologies to better understand the lung microbiome is a promising area of research that may offer further insight into COPD.
Declaration of Interests
Dr. Martinez has been a consultant for Amgen, Carden Jennings, CSA Medical, Forest, Ikaria, Jansen, Merck, Mycomed/Takeda, GlaxoSmithKline, Gilead Sciences, Promedior, Bayer, Johnson & Johnson. He has served as a committee member for Forest, Jassen, Nycomed/Takeda, GlaxoSmithKline, Gilead Sicencs, Promedior, Bayer, Johnson & Johnson and served on the Food and Drug Administration Mock Panel for Boehringer Ingelheim and GlaxoSmithKline He has served on the Data Safety and Monitoring Boards of Stromedix and GlaxoSmithKline. He has been a speaker for Nycomed/Takeda and participated in CME programs for Annenberg, CME Incite, MedScape, Miller, Paradigm, Peer Voice and NACE.