Running Head: Subtyping COPD with Blood Proteomics
Funding Support: Supported by National Institutes of Health grants R01HL094635, P01HL105339, R01HL125583, R01HL130512. The TESRA trial was supported by Roche.
Date of Acceptance: November 1, 2016
Abbreviations: chronic obstructive pulmonary disease, COPD; Treatment of Emphysema with a gamma-Selective Retinoid Agonist trial, TESRA; computed tomography, CT; Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints, ECLIPSE; forced volume in 1 second, FEV1; forced vital capacity, FVC; diffusing capacity of the lungs for carbon monoxide, DLCO; St. George’s Respiratory Questionnaire, SGRQ; 15th percentile of lung density, Perc 15; Hounsfield units, HU; ethylenediamine-tetraacetic acid, EDTA; analysis of variance, ANOVA; Database for Annotation, Visualization and Integrated Discovery, DAVID; matrix metalloproteinase 9, MMP9; transforming growth factor beta, TGF-β; C-reactive protein, CRP; pulmonary and activation-regulated chemokine, PARC; chemokine ligand 18, CCL18; interleukin-18, IL-18; brain-derived neurotrophic factor, BDNF; platelet-derived growth factor, PDGF; epidermal growth factor, EGF; vascular endothelial growth factor, VEGF; CC chemokine ligand 16, CCL16; interleukin-1, IL-1; interleukin-8, IL-8; interleukin-10, IL-10; Body mass index-airflow Obstruction-Dyspnea-Exercise capacity index, BODE; modified Medical Research Council dyspnea index, MMRC
Citation: Zarei S, Mirtar A, Morrow JD, Castaldi PJ, Belloni P, Hersh CP. Subtyping chronic obstructive pulmonary disease using peripheral blood proteomics. Chronic Obstr Pulm Dis. 2017; 4(2): 97-108. doi: http://doi.org/10.15326/jcopdf.4.2.2016.0147
Introduction
Chronic obstructive pulmonary disease (COPD) is a common, progressive disease defined by airflow limitation on lung function tests. COPD is a heterogeneous condition, characterized by varying symptoms, natural history, and anatomic processes, which can be visualized on chest computed tomography (CT) scans.1 These differences may be attributable to the molecular heterogeneity of the disease.2 Starting almost 40 years ago, researchers have attempted to define the spectrum of heterogeneity in COPD.3 However the underlying pathogenesis for the heterogeneity of the disease is still under investigation. Hence, identifying COPD phenotypes with the goal of achieving individualized treatment remains an important goal.4 To date, studies have identified several clinical subtypes of COPD as well as a genetic subtype, alpha-1 antitrypsin deficiency, that can be targeted with specific treatments.5-7
Statistical techniques such as cluster analysis have been used to assign COPD individuals to different groups where individuals within each cluster have more common characteristics compared to individuals from other groups or clusters.4 These techniques have relied upon clinical and physiologic variables, chest CT scans, and gene expression data.4,8,9 A recent COPD subtyping analysis in the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) study used a limited set of blood biomarkers, including club cell secretory protein-16, surfactant protein-D, interleukin-8, tumor necrosis factor, fibrinogen and white blood cell counts.10 The latter 2 biomarkers may be the most clinically applicable.10 However, none of the previous studies have used large-scale proteomics to find different subgroups of COPD. In this study, we analyzed a large set of peripheral blood biomarkers in former smokers with emphysema from a clinical trial. We hypothesized that unbiased mathematical tools such as hierarchical clustering will be able to define sets of biomarkers that delineate the clinical variation among COPD patients and provide insight into the potential pathogenic mechanisms behind the subgroups.
Methods
Study Participants
The Treatment of Emphysema with a Selective Retinoid Agonist (TESRA) trial was a multicenter randomized, placebo-controlled clinical trial of palovarotene in COPD (clinicaltrials.gov identifier NCT00413205).11 Details on the design, methods and the collection of clinical data of the TESRA study have been previously described.11,12 Study participants provided written informed consent, and the study was approved by the institutional review boards at all participating centers. The study enrolled former smokers (abstinent at least 12 months) with a minimum of 10 pack years smoking history. Study participants had moderate-to-severe COPD, defined by post-bronchodilator forced expiratory volume in 1 second (FEV1) to forced vital capacity (FVC) ratio <0.7 and FEV1 < 70 % predicted, with diffusing capacity of the lung for carbon dioxide (DLCO) < 70% predicted. All participants had emphysema based on visual review of chest CT scans. Baseline measurements also included assessments of symptoms and exacerbations, a 6-minute walk test, and body plethysmography to measure lung volumes. Disease-related quality of life was assessed using the St. George’s Respiratory Questionnaire (SGRQ).13 Chronic bronchitis was defined using the SGRQ per Kim et al.14 Exacerbations were identified by hospitalization or treatment with oral steroids or antibiotics. Emphysema was quantified by the 15th percentile (Perc15) of lung density histogram .15 Since Perc15 values are negative, the variable was converted to a positive value before log-transformation i.e., Perc15-transformed = log10 (1000 + Perc15). Therefore, higher values of the transformed variable correspond to lower quantitative emphysema. The percent of voxels with attenuation < -910 Hounsfield units (HU) was also used to assess emphysema.
Biomarker Measurements
Prior to randomization, blood samples were collected from 458 participants for biomarker analysis. These biomarkers were selected based on presumed biologic mechanisms in COPD.12 To measure the biomarker levels, a custom 15-panel assay was used. Concentrations of 140 protein biomarkers were measured in ethylenediamine-tetraacetic acid (EDTA) plasma in duplicate at Rules Based Medicine (Austin, Texas) and Quest Diagnostics (Valencia, California). Full details of the biomarker testing in TESRA, including the list of biomarkers, have been previously published.12,16 For this analysis, biomarker measurements below the lower limit of quantification were set to missing. Otherwise, untransformed biomarker values were used.
Cluster Analysis
We implemented a simple optimization (maximization) algorithm to include the maximum number of participants and biomarkers, while minimizing missing data. Details of the algorithm can be found in the online supplement, Supplemental Figure 1. The maximum amount of data without missing values is found with 87 biomarkers and 396 participants. We performed an agglomerative McQuitty hierarchical clustering using only the biomarker dataset with a Canberra distance method to identify participant clusters on the basis of their individual biomarker profiles.17 Canberra distance is suitable for biomarker data which contains large values, outliers and non-normally distributed variables.18 To evaluate the quality of clusters and to find the optimal number of clusters, the R package NbClust was used.19 NbClust provides multiple indices which determine the optimal number of clusters in a dataset. We used analysis of variance (ANOVA) to compare the mean values of phenotypes and biomarkers across different clusters. When the ANOVA p-value was significant, Tukey’s test was used for pairwise comparisons between the groups.
Enrichment Analysis
For the biomarkers that had different mean values among subgroups of COPD, we utilized enrichment analysis to find the corresponding molecular pathways for these biomarkers using Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.720 and Reactome gene enrichment tools.21 The biomarker proteins were mapped to their corresponding genes. Gene lists were input into GeneMANIA to create graphical networks.22 GeneMANIA draws edges between the query genes and extends the set of genes based on databases of genetic interactions, molecular pathways, co-expression, co-localization, physical interactions and shared protein domains.
Results
Based on the majority (12 of 23) of the metrics implemented in the NbClust package,19 we determined the optimal number of clusters to be 3 (online supplement, Supplemental Table 1). Therefore, we used hierarchical clustering of the biomarker measurements to divide the COPD participants into 3 subgroups, containing 267 (67.4%), 104 (26.3%) and 25 (6.3%) participants (Figure 1, Supplemental Figure 2).
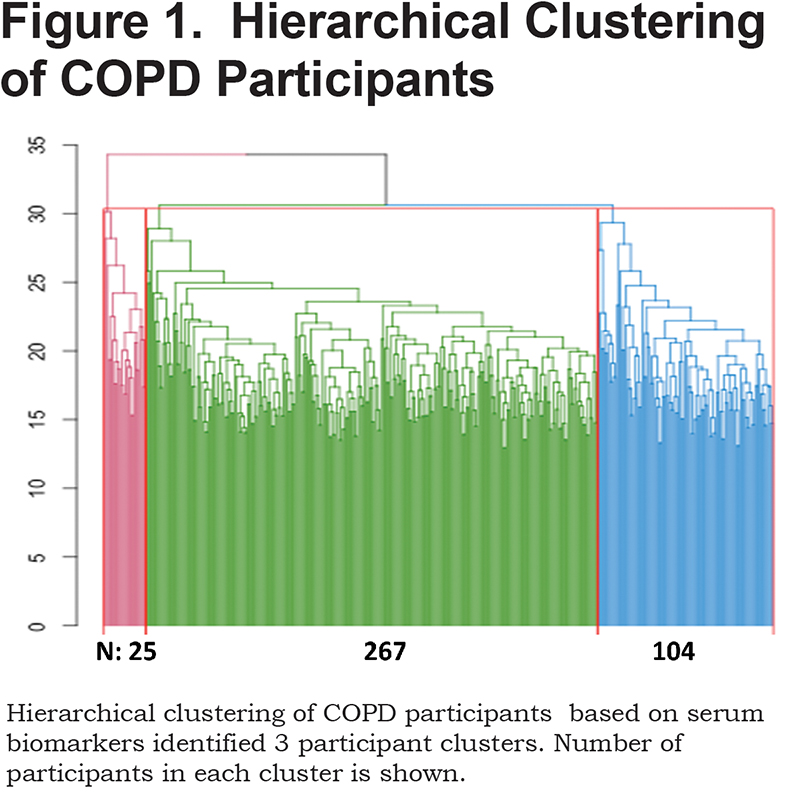
The characteristics of participants in each cluster are listed in Table 1. According to the ANOVA, the mean values for emphysema, measured by the 15th percentile of lung density histogram (transformed, see Methods), as well as the total score on the SGRQ were different between the 3 groups. Participants in the third cluster had higher SGRQ scores, consistent with a lower quality of life. However, they had less emphysema on chest CT scans (higher transformed Perc15 values). Cluster 3 showed a trend for the lowest emphysema, based on untransformed Perc15 and -910HU threshold, but the difference between clusters was not statistically significant. SGRQ scores and emphysema were not significantly different between clusters 1 and 2.
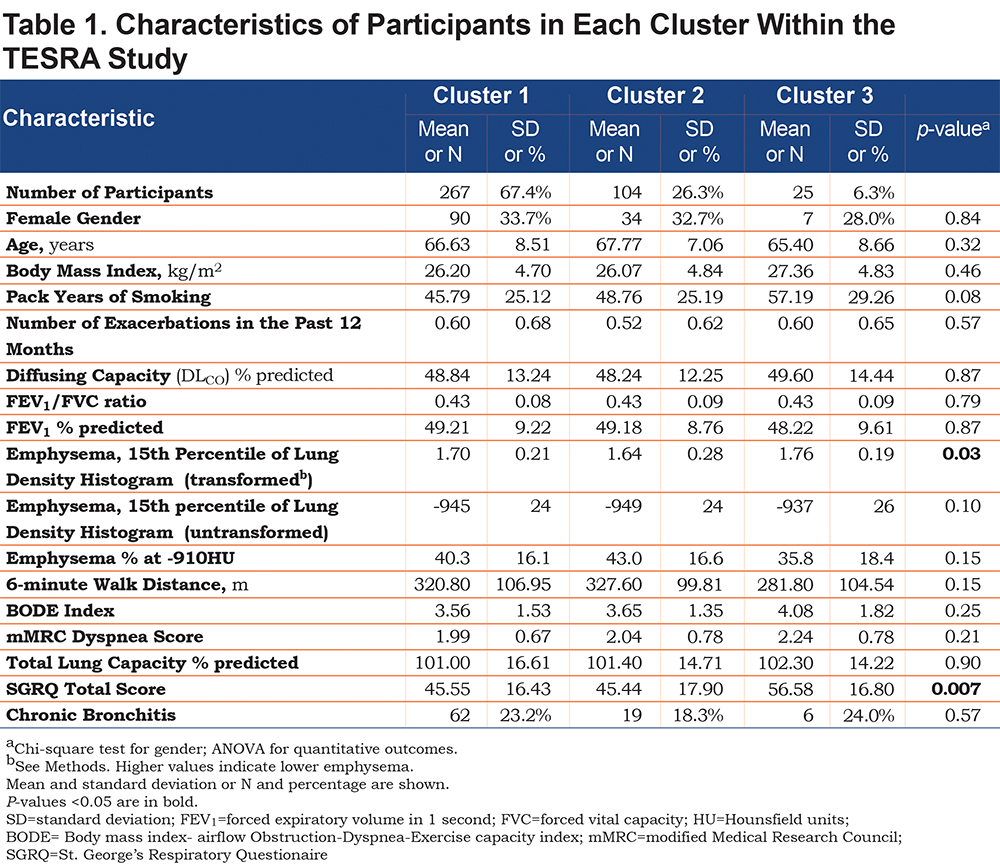
Table 2 lists the 18 biomarkers that are significantly different between the 3 clusters, based on ANOVA p-value<0.05 and p-values<0.05 for all pairwise Tukey tests. Several of these proteins are known to be important in COPD pathogenesis, such as matrix metalloproteinase 9 (MMP9) and transforming growth factor beta (TGF-β). 23,24 Many of the biomarkers remained significant after Bonferroni correction for multiple testing (p-value<0.05/87=5.7e-4). The biomarkers in Table 2 were only weakly correlated with pack years of smoking (absolute value of Pearson r ≤0.2).
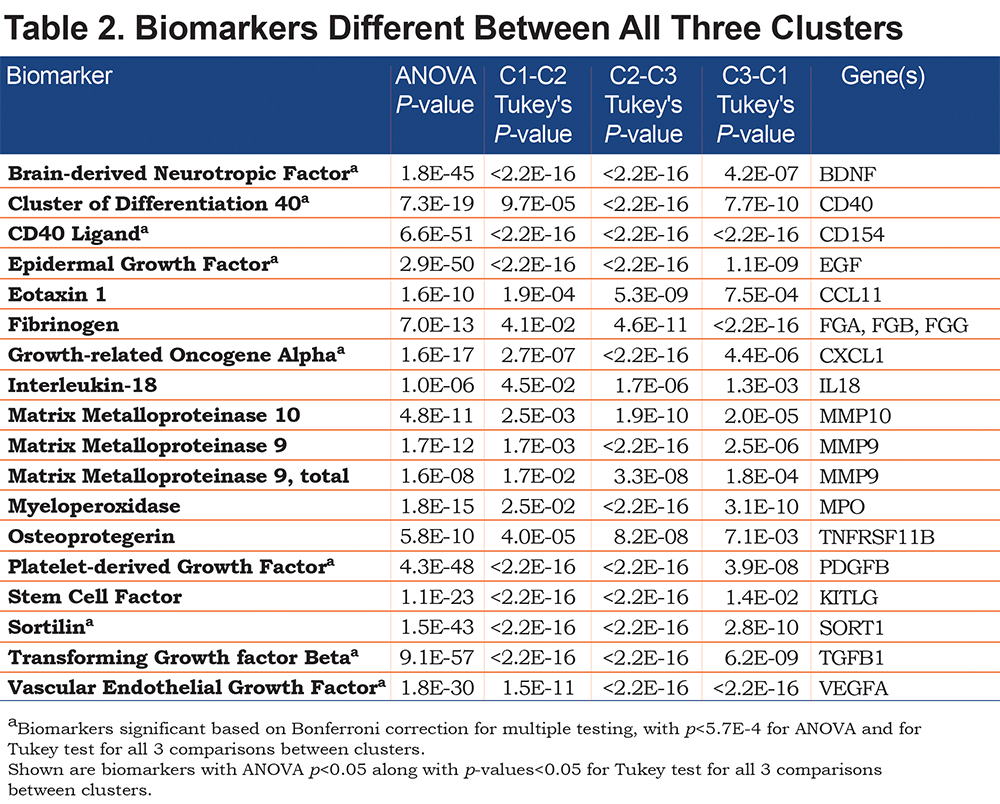
Enrichment analysis using DAVID, GeneMANIA and Reactome was performed using the list of genes coding for the biomarkers in Table 2. Figure 2 illustrates the gene and pathway enrichment analysis methods used, and the resulting top enriched pathways and the relevant genes. The Table 2 biomarkers were enriched for platelet-related pathways.
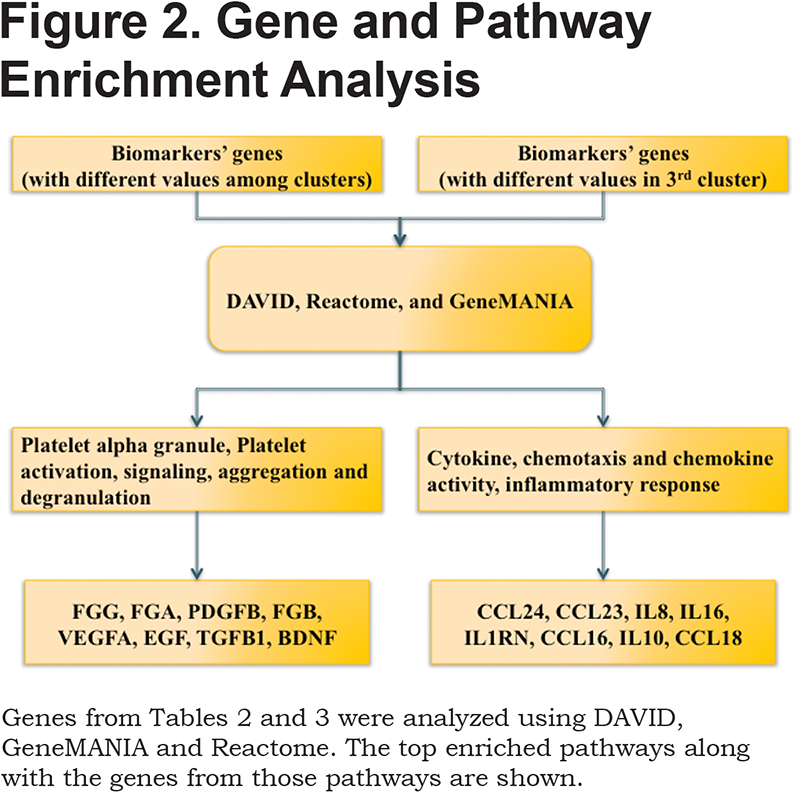
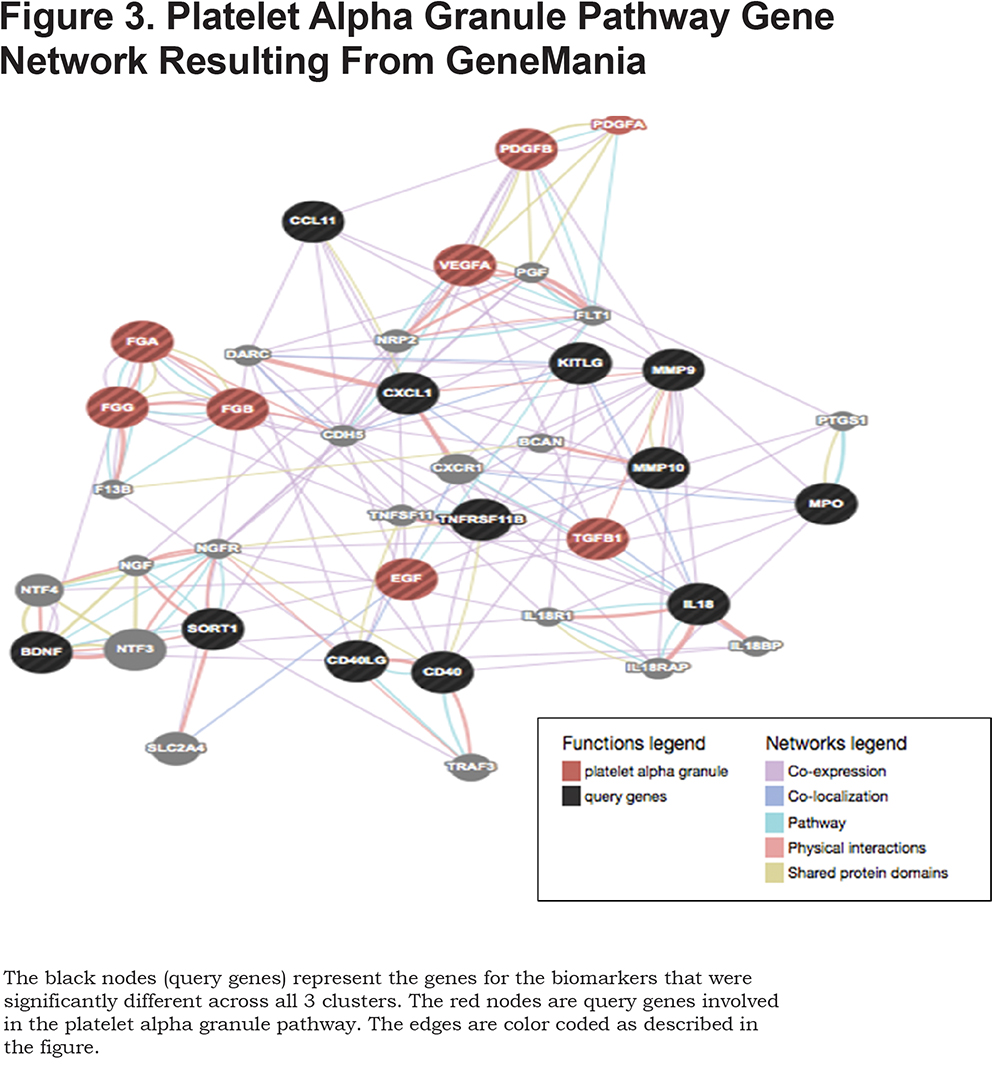
Supplemental Table 2 displays the result of DAVID enrichment analysis for the genes coding for the biomarkers listed in Table 2, limited to results with false discovery rate < 0.05. Pathways are presented in annotation clusters with similar mechanisms and similar genes. The second annotation cluster included platelet granule genes, which are listed in Figure 2. Similar results were seen with GeneMANIA and Reactome. The GeneMANIA network of genes involved in platelet alpha granule pathway is displayed in Figure 3. The identified biomarker genes have multiple network connections in this pathway.
Based on the phenotype differences in cluster 3 seen in Table 1, we focused on biomarkers that showed different values in the participants from the third cluster compared with each of the first 2 clusters; these 21 biomarkers are listed in Table 3. Several of these biomarkers have been linked to COPD in previous studies. C-reactive protein (CRP), pulmonary and activation-regulated chemokine (PARC)/chemokine ligand 18 (CCL18), and interleukin (IL)-18 are known inflammatory biomarkers relevant to COPD25-27; all had higher levels in cluster 3. Alpha-1 antitrypsin levels were higher in cluster 3, consistent with the lower quantitative emphysema values in these participants. Similar to Table 2, the biomarkers in Table 3 were weakly correlated with pack years (absolute value of Pearson r ≤0.2).
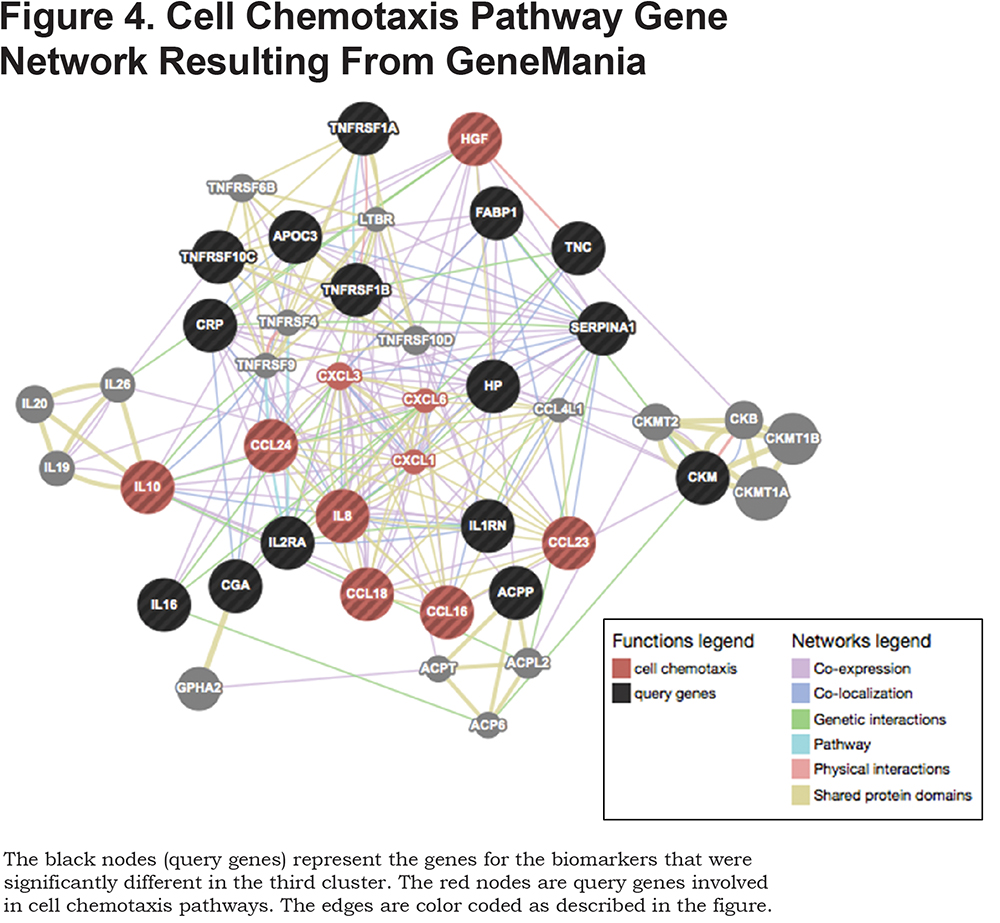
Supplemental Table 3 shows the DAVID enrichment analysis for the genes encoding the biomarkers in Table 3. The top 2 annotation clusters relate to cytokines and chemotaxis, which was confirmed in GeneMANIA and Reactome. The GeneMANIA network analysis of these corresponding genes involved in cytokine/chemotaxis activities is displayed in Figure 4.
Supplemental Tables 4 and 5 show the mean levels by cluster for those biomarkers in the platelet alpha granule and cytokine/chemotaxis annotations, respectively. Participants from the first cluster have the lowest values of alpha platelet granule biomarkers while participants from the third cluster have the highest values. Participants from the third cluster have the highest values for the chemotaxis-related biomarkers.
Discussion
In a study of former smokers with moderate-to-severe COPD with emphysema on chest CT scans, we were able to identify 3 subgroups based on cluster analysis of blood proteomics data. We specifically defined a small subgroup of participants with increased inflammatory biomarkers, yet less emphysema on quantitative analysis of chest CT scans, based on the 15th percentile of lung density histogram, which may be a better measure of emphysema in COPD than the -910HU threshold.28 These participants had similar reductions in lung function, suggesting that airway disease may be present as well. This subgroup had reduced disease-related quality of life, exceeding the minimum clinically important difference of 4 points in total SGRQ score.29 A previous study had used a panel of blood and sputum biomarkers to define 4 biologic clusters of acute exacerbations,30 but our study is the first to use proteomics to subtype stable COPD patients.
Gene enrichment analysis showed that biomarkers annotated to the platelet alpha granule pathway were different between the 3 subgroups of COPD. Platelets contain different storage granules including alpha granules, dense granules and lysosomes. Alpha granules, the main storage granules, contain fibrinogen, von Willebrand factor, growth factors and protease inhibitors that enhance thrombin formation at the site of injury.31 Several of the platelet alpha granule biomarkers have previously been shown to be associated with COPD and related phenotypes. Systemic inflammatory markers such as fibrinogen are associated with COPD risk, mortality and exacerbations.32,33 Brain-derived neurotrophic factor (BDNF) is stored in platelets and is released during an inflammatory response.34 BDNF has been shown to be elevated in COPD individuals when compared to controls.34,35 Growth factors, including platelet-derived growth factor (PDGF), epidermal growth factor (EGF), and TGF-β, are mitogens for smooth muscle cells and fibroblasts. PDGF may be involved in vascular and small airway remodeling in COPD.36,37 EGF and TGF-β are both upregulated in airway epithelium and submucosal cells of patients with COPD.38,39 The role of these growth factors, especially TGF-β, in airway remodeling in COPD has been well-documented.23,40-42
The important pro-angiogenic, regulatory protein, vascular endothelial growth factor (VEGF), is also housed in alpha granules. VEGF plays a role in maintaining the homeostasis of alveoli; decrease in VEGF expression is associated with pulmonary endothelial cell apoptosis.43,44 In our study, participants from the third cluster have the highest mean values for the platelet alpha granule biomarkers, including VEGF, and the lowest amount of emphysema on chest CT scans.
In the analysis focusing on differences between the third cluster and the other 2 clusters, gene enrichment pointed to genes related to cell chemotaxis and inflammatory response. The corresponding biomarkers included PARC, CC chemokine ligand 16 (CCL16), interleukin-1 (IL-1) receptor antagonist, interleukin-8 (IL-8) and interleukin-10 (IL-10). PARC/CCL-18is primarily synthesized in dendritic cells and monocytes and is highly expressed in the lungs.45,46 Serum PARC/CCL-18 is elevated in COPD and is associated with mortality.25
Balanced secretion of pro- and anti-inflammatory cytokines is essential in limiting pulmonary inflammation in the stable state and during respiratory infections. IL-10, an anti-inflammatorycytokine, was shown to be decreased in serum and sputum of COPD participants and healthy smokers compared to non-smokers.47 On the other hand, pro-inflammatory cytokines such as IL‐8, secreted by alveolar macrophages, were found to be elevated in COPD patients and were further increased during exacerbations.40 On average, participants from the third cluster had the highest mean values for both pro- and anti-inflammatory biomarkers.
Our study has several limitations. The biomarker panel used was selected from available assays, based on possible mechanisms in COPD. The panel was heavily weighted towards inflammatory markers, which may explain why these pathways were prominent in our results. Unbiased proteomic assays would be required to identify novel pathways in COPD subgroups. Based on enrollment criteria in the TESRA trial, which included moderate-to-severe COPD with emphysema, study participants tended to be more homogeneous than the general COPD patient population, which may limit the ability to identify subgroups (as demonstrated in Supplemental Figure 2) and may also limit generalizability. Despite these limitations, we were able to identify 3 subgroups of COPD participants using clustering and network analysis of a large panel of serum biomarkers. We found participants in the smallest subgroup to have the highest levels of platelet alpha granule biomarkers and inflammatory cytokines. These participants had less emphysema and a lower quality of life, despite similar levels of lung function impairment. Thus, these individuals may have more airway inflammation compared to the other groups. Future studies measuring similar biomarkers in a broader range of COPD participants will be required to validate and to expand upon these results. The ultimate goal is to use serum biomarkers to define clinically-relevant COPD subgroups which may have different outcomes and could potentially be treated with different therapies.
Acknowledgements
TESRA Investigators: Ognian Georgiev, Dimitar Popov, and Vasil Dimitrov, Sofia, Bulgaria;Hristo Metev, Ruse, Bulgaria; Yavor Ivanov, St. Pleven, Bulgaria; Libor Fila and Jiri Votruba, Praha, Czech Republic; Vladimir Zindr Vitezna, Karlovy Vary, Czech Republic; Kamil Klenha, Tabor, Czech Republic; Jaromir Roubec, Ostrava, Czech Republic; Barna Szima, Szombathely, Hungary; Zsuzsanna Mark, Torokbalint, Hungary; Zoltan Baliko, Pecs Hungary; Zoltan Bartfai, Budapest, Hungary; Katalin Gomori, Balassagyarmat, Hungary; Andres Sigvaldason, Reykjavik, Iceland; Mordechai Kramer, Petach Tikva, Israel; Gershon Ya Fink, Rehovot, Israel; Zeev Weiler, Ashkelon, Israel; Joel Greif, Tel Aviv, Israel; Issahar Ben-Dov, Ramat Gan, Israel; Mordechai Yigla, Haifa, Israel; Leonardo Fabbri, Modena, Italy; Pierluigi Paggiaro, Pisa, Italy; Giorgio Canonica, Genova, Italy; Isa Cerveri, Pavia, Italy; Antra Bekere and Aurika Babjoniseva, Riga, Latvia; Alvil Krams and Stopinu Pagasts, Rigas Rajons, Latvia; Wladyslaw Pierzchala, Katowice, Poland; Dariusz Nowak, Lodz, Poland; Robert Mroz, Bialystok, Poland; Hanna Szelerska-Twardosz and Malgorzata Rzymkowska, Poznan, Poland; Ismail Abdullah, Durban, South Africa; Christo Van Dyk, Western Cape, Worcester, South Africa; Nyda Fourie, Bloemfontein, South Africa; John O’Brien and Mary Batema, Cape Town, South Africa; J. Joubert, Bellville-Cape Province, South Africa; Abdool Gafar, Kwa-Zulu Natal, Amanzimtoti, South Africa; Hannes Van Rensburg, Centurion, South Africa; Lyudmila Yashina, Oleksandr Dzyublik, Volodymyr Gavrysyuk and Yuriy Feshchenko, Kiev, Ukraine; Nadezda Monogarova, Donetsk, Ukraine; David Parr, Coventry, United Kingdom; Stephen Rennard, Omaha, Nebraska; Richard Casaburi, Torrance, California; Gerard Criner, Philadelphia, Pennsylvania; Mark Dransfield, Birmingham, Alabama; Charles Fogarty, Spartanburg, South Carolina; Nicola Hanania and Amir Harafkhaneh, Houston, Texas; Carl Griffin and Kathi Mcdavid, Oklahoma City, Oklahoma;, Paul Kvale, Detroit, Michigan; Barry Make, Denver, Colorado; Joe Ramsdell, San Diego, California; Michael D. Roth, Los Angeles, California; Peter Sporn, Chicago, Illionois.
Steering Committee: Alvar Agustı´ (Spain), Peter Calverley (United Kingdom), Leonardo Fabbri (Italy), Klaus F. Rabe (Netherlands), Nicolas Roche (France), Michael Roth (United States), Jorgen Vestbo (Denmark), Stephen Rennard (United States).
Declaration of of Interest
Dr. Hersh reports consulting fees from AstraZeneca, Concert Pharmaceuticals and Mylan. Dr. Belloni is an employee of Genentech. Drs. Zarei, Mirtar, Morrow, and Castaldi report no competing interests. Roche designed the TESRA trial and collected the data. The funders had no role in the data analysis in this manuscript, the writing of the manuscript or the decision to submit the manuscript for publication.