Running Head: Respiratory Pathogens in Small Airways in Stable COPD
Funding Support: AstraZeneca and the Canadian Institutes of Health Research.
Date of Acceptance: November 30, 2022 | Published Online Date: December 2, 2022.
Abbreviations: BAL=bronchoalveolar lavage; COPD=chronic obstructive pulmonary disease; FEV1=forced expiratory volume in 1 second; PCR=polymerase chain reaction; PPB=potentially-pathogenic bacteria; SGRQ=St George’s Respiratory Questionnaire
Citation: Alotaibi NM, Tam S, van Eeden SF, et al. Common respiratory pathogens other than haemophilus in small airways are associated with neutrophilic inflammation and poor health status in stable COPD patients. Chronic Obstr Pulm Dis. 2023; 10(1): 122-126. doi: http://doi.org/10.15326/jcopdf.2022.0338
Online Supplemental Material: Read Online Supplemental Material (238KB)
Introduction
Chronic obstructive pulmonary disease (COPD) is an inflammatory disease characterized by a predominantly neutrophilic pattern of inflammation, which persists even after smoking cessation; inflammation in the small airways is believed to be a key mechanism for COPD progression and exacerbations.1 It has been observed that the presence of potentially-pathogenic bacteria (PPB) identified via culture is associated with neutrophilic airway inflammation and is a possible driver of disease progression.2 Using a multiplex polymerase chain reaction (PCR) pathogen detection array, we determined the presence of potentially-pathogenic microorganisms in the bronchoalveolar lavage (BAL) fluid of stable COPD patients and evaluated its relationship with airway inflammation in BAL fluid and health status of patients as measured by the St George’s Respiratory Questionnaire (SGRQ).
Methods
After receipt of informed consent, we performed flexible bronchoscopy and collected BAL samples from the first 38 participants with stable COPD who participated in the Study to Investigate the Differential Effects of Inhaled Symbicort and Advair on Lung Microbiota (DISARM) study (clinicaltrials.gov identifier NCT02833480) which received institutional ethics approval from the University of British Columbia and the Providence Health Care Research Ethics Committee (H14-02277). Details of the DISARM study have been published previously.3 Briefly, all participants were free of exacerbations for at least 8 weeks and did not take any inhaled corticosteroids for at least 4 weeks prior to bronchoscopy. During these 4 weeks, all participants were treated with inhaled formoterol (12ug twice daily; Oxeze Turbuhaler®). Immediately before bronchoscopy, all patients rinsed their mouths with 10 ml of sterile saline, followed by an antiseptic wash with 5ml of Listerine®. Following conscious sedation, a fiberoptic bronchoscope was inserted into the patients’ oropharynx. Using no or minimal suction, the bronchoscope was introduced into the lower airways and wedged into the smallest airway possible in the right middle or upper lobe; 2% lidocaine was used to anesthetize the airways. Approximately 100ml–200ml of normal saline divided into 3–5 aliquots were instilled into the wedged airway from which at least 10ml of fluid was recovered (Table 5 in the online supplement which includes the volume of BAL return for each participant). The recovered fluid was centrifuged at a speed of 250 × g for 10 minutes at 4°C and the cellular components were resuspended in Dulbecco's phosphate buffered saline without Ca++ and Mg++. The resuspended pellet was then split into 2 tubes and centrifuged at a speed of 300 × g for 5 minutes at 4°C. Next, the supernatant was discarded and 1 of the 2 pellets was resuspended in 900ul Cytolyt solution and stored at -80°C. We used 200ul of the BAL pellet resuspended in Cytolyt in this study; nucleic acids were extracted using QIAamp MinElute Virus Spin Kit (Qiagen) according to manufacturer’s instructions. The extracted nucleic acids were tested on the Randox® cystic fibrosis respiratory pathogen panel, which interrogates 21 bacteria, 7 viruses, and 4 fungi that are considered to be common potentially-pathogenic organisms in the airways (Figure 1). The panel employs multiplex PCR and biochip array hybridization technology. Total cell count on the BAL fluid was measured using hemocytometer, and cell differentials were measured via cytocentrifugation in accordance with the American Thoracic Society guidelines.4
Results
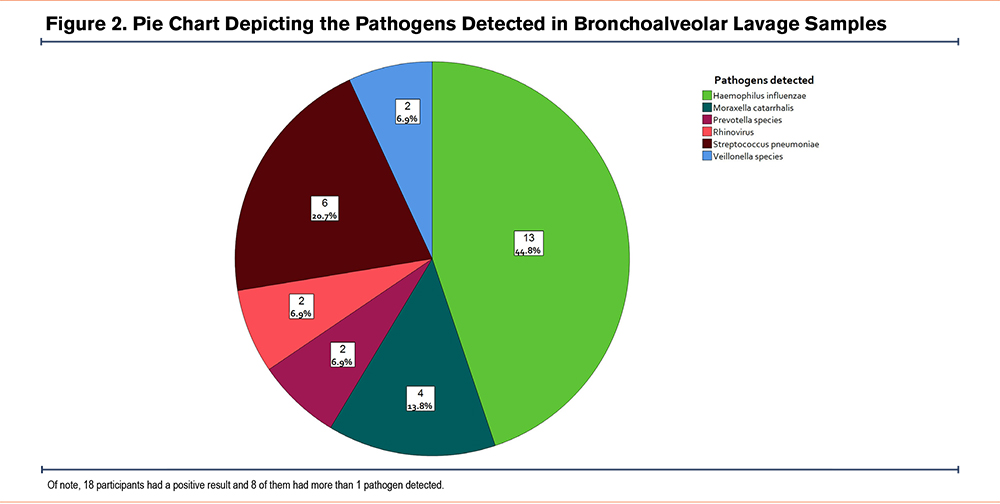
The participants had a mean age of 64±8 years, mean forced expiratory volume in 1 second (FEV1) of 57±16 percentage of predicted, and a mean total SGRQ score of 43.18±15.84 units; 89.4% were men, and 44.7% were current smokers (the rest were ex-smokers with at least a 10 pack-year history). Molecular pathogen testing demonstrated that 18 participants (47.4%) had pathogens detected on their BAL cell pellets while the rest had a negative result (Figure 2). Participants with pathogens detected had significantly higher BAL-neutrophil% (pathogen-positive group, median 20% interquartile range [5.6% –45.2%] versus the pathogen-negative group, 4% [2%–16.2%]; P=0.024; online supplement Table 6). There was no statistically significant difference in SGRQ scores between the 2 groups (pathogen-positive mean ±SD, 47.4±16.4 versus pathogen-negative, 39.6±14.7; P=0.136).
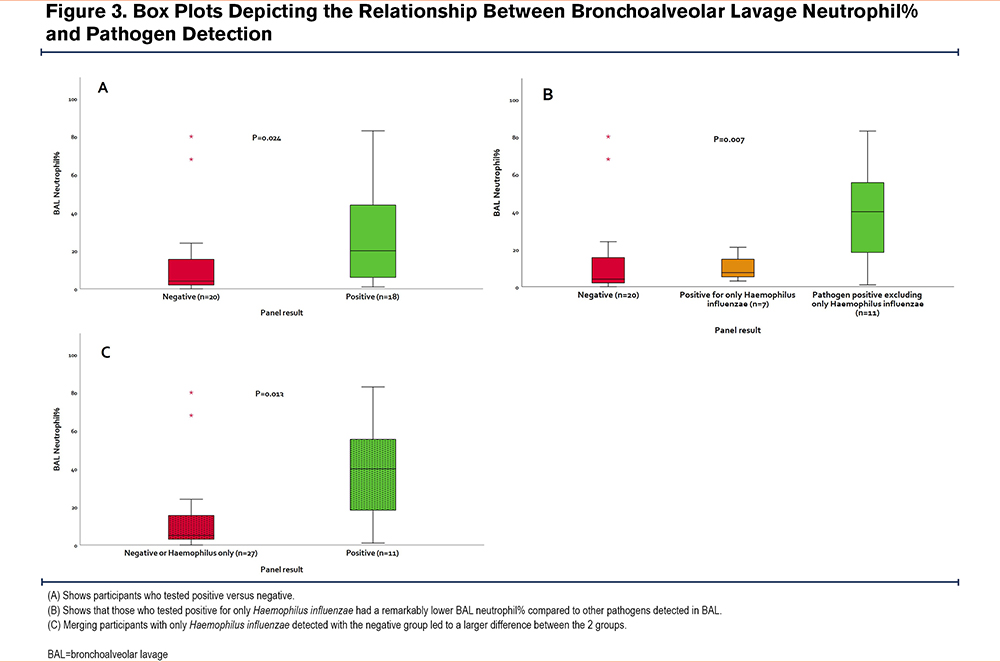
Seven participants had a positive result for only Haemophilus influenzae on BAL. These participants had significantly lower BAL neutrophil% compared to the other participants who were pathogen-positive (Haemophilus-only, 7.5% [4.5%–19%] versus pathogen-positive, 40% [8%-62%]; P=0.007) (Figure 3). As Haemophilus could be a colonizer (rather than a pathogen), we merged the pathogen-negative group (n=20) with those who only tested positive for Haemophilus (n=7). The pathogen-positive exclusive of Haemophilus group (n=11) had significantly higher BAL-neutrophil% compared with the pathogen-negative or the Haemophilus-only group (pathogen-positive, 40% [8%–62%] versus pathogen-negative or Haemophilus-only, 5% [3%–17%]; P=0.013; Figure 3; online supplement Table 7). On multivariate regression controlling for age, sex, FEV1% of predicted and smoking status, pathogen-positivity exclusive of Haemophilus-only was independently associated with BAL neutrophil% (P=0.003).
Total SGRQ score was also higher (pathogen-positive, 51.3±18 versus pathogen-negative or Haemophilus-only, 39.7±13.7; P=0.039). While the domains of SGRQ demonstrated similar results, only the activity domain showed statistical significance (pathogen-positive, 70.8±17.6 versus pathogen-negative or Haemophilus-only, 58.6±15.2; P=0.039).
Discussion
To our knowledge, this is the first study of its kind to examine cell pellets from BAL fluid in stable COPD individuals for potential airway pathogens using a molecular pathogen detection method that is commercially available. We found that approximately 50% of patients (not on inhaled corticosteroids and free of exacerbations) demonstrated a positive result on molecular testing, which in turn, was associated with intense neutrophilic inflammation in the small airways and with poor health status. The only exception was the presence of Haemophilus influenzae,which was not associated with airway neutrophilia.
Haemophilus was the most commonly detected organism in our study. Importantly, we observed that participants who had Haemophilus-only did not demonstrate significant neutrophilic inflammation. We speculate that in stable COPD patients, Haemophilus, unlike other organisms on the panel, is a colonizer rather than a pathogen and as such does not elicit a strong innate immune response in the host. This notion is consistent with prior observations that suggest Haemophilus is uniquely adapted to establish colonization in COPD.5 Although the exact mechanisms by which Haemophilus colonizes the airways in COPD have not been fully elucidated, there is evidence to suggest that Haemophilus influenzae could be resistant to neutrophil extracellular traps and thus, avoid neutrophil-mediated killing.6 Additional studies will be required to unravel the exact mechanisms.
Our results are consistent with prior studies that showed the presence of PPB in the sputum of COPD patients was associated with worsening of daily symptoms and increased airway inflammation.7 We extend these findings by demonstrating pathogen-positivity in BAL cell pellets exclusive of Haemophilus-only was associated with worse health status in patients free of exacerbations and inhaled corticosteroids. These data raise the possibility that these microbes may contribute to persistent (neutrophilic) inflammation in the small airways, which in turn, may lead to poor health status of patients. Poor health status has been associated with increased risk of exacerbations and accelerated decline in lung function.8,9 Additional studies will be required to determine whether treating these microbes or the associated neutrophilic inflammation can ameliorate health outcomes of patients.
A limitation of our study is that we had a relatively small sample size that impeded our ability to examine pathogens other than Haemophilus independently. Moreover, the panel that we used is qualitative in nature and not quantitative, thus, dose-response relationships could not be ascertained. We also cannot definitively rule out the possibility of contamination as a cause of the positive result in some participants. However, we have shown previously, using the same samples, that bacteria abundance as measured on 16S sequencing is extremely low in bronchial channel washes and extraction negative controls,3 which renders contamination as the primary driver of our analysis unlikely. We did not find a significant association between DNA concentration as a metric of sample quality or BAL return volumes and the probability of testing positive on the multiplex array (online supplement Table 5).
In conclusion, we demonstrate here that the presence of potentially-pathogenic microorganisms (other than Haemophilus influenzae) in stable COPD is associated with intense neutrophilic inflammation and poor health status. Targeting of anti-inflammatory and/or anti-microbial therapies in these patients may lead to better outcomes in the future.
Acknowledgements
Author Contributions: All authors contributed to the study conception and design of the work. DDS, JML, SFvE, TS, and SL performed the research bronchoscopies. NMA performed nucleic acid extractions, data analysis, and drafting of the initial manuscript. All authors participated in the interpretation of the data and have read, revised, and approved the final manuscript.
Declaration of Interest
DDS has received honorarium for COPD talks from AstraZeneca, GlaxoSmithKline, and Boehringer Ingelheim. He is also a Tier 1 Canada Research Chair and holds the de Lazzari Family Chair at the Heart Lung Innovation Centre. JML is a Tier 2 Canada Research Chair in Translational Airway Biology. All other authors have nothing to disclose.